Davis, M. P., Sparks, J. S., & Smith, W. L. (2016). Repeated and Widespread Evolution of Bioluminescence in Marine Fishes. Plos One, 11(6), e0155154. doi:10.1371/journal.pone.0155154
“A light from the shadows shall spring”
In light-limited environments, like the deep sea, organisms will inevitably adapt in unique and mind-boggling ways in an effort to communicate, to find prey, to defend themselves, etc. One such adaptation to counteract the dark environment is by developing bioluminescent capabilities—that is, the ability for animals or bacteria within them to produce light. Depending on where you grew up, you may have encountered fireflies (lightning bugs) in your youth. These insects are an excellent example of harnessing light for communication as their abdomens blink in the fading light of a summer night to find mates. But fireflies aren’t alone on the tree of life when it comes to glowing in the dark. In fact, bioluminescence evolved over forty separate times throughout the animal kingdom—or so we thought until Matthew Davis and his team began looking at the phenomenon in fishes.
The evolution of bioluminescence in fishes used to be counted as a singular event; however, the intricate nature of the bioluminescent process suggests light production must have emerged separately within different groups of fishes. Bioluminescence can be intrinsic—that is, the fish produces the substances necessary for light production itself—or bacterially-mediated—when the fish hosts the bacteria responsible for the chemical reaction that produces light. To that end, many things have to align for the ability to even evolve this ability. But how would one go about determining how unique each appearance of bioluminescence is, or when it first appeared in a group of fishes? In this case, it comes down to the genes.
Building a Phylogeny
Davis and his team started constructing an evolutionary tree based on similarities and differences in genetic characters associated with bioluminescence. They used eleven distinct gene fragments collected from nearly 300 individual genera of fishes while building on previous phylogenies to create a new tree. Using computer modeling and statistics, the team constructed a tree with the greatest likelihood of correctly depicting all instances of bioluminescence when they independently evolved.
Illuminating Results
Surprisingly, Davis and his colleagues determined that bioluminescence has evolved independently twenty-seven times across bony fish families (Fig. 1). When adding in other work done on bioluminescence in sharks (cartilaginous fish), the total tally of independent events comes in at twenty-nine occurrences. Marine fishes are the only known vertebrates to have developed bioluminescence, and they’ve done so more often than expected.

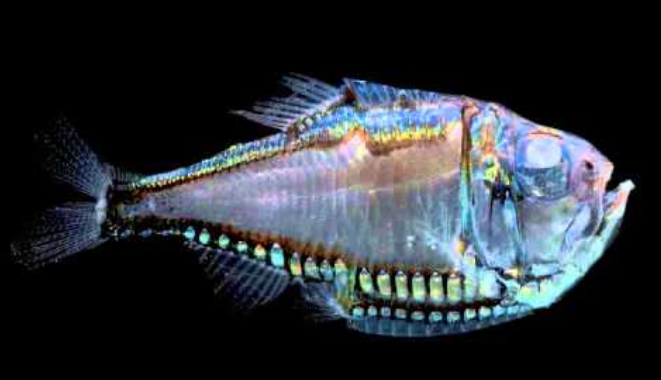
With respect to the types of bioluminescence, Davis found that the bacterially-mediated type emerged seventeen distinct times, although the type of bacteria remained the same regardless of the species of host fish. Many of these groups of fishes have also evolved unique physical adaptations to further utilize their light-producing abilities. For example, anglerfishes (Fig. 2A) have illuminated lures atop their heads, presumably to attract prey while ponyfishes (Fig. 2B) have a type of muscular curtain that can be drawn over pockets of glowing bacteria to limit the amount of light escaping. Intrinsic bioluminescence evolved less often: a mere eight times. However, for as little as it evolved, intrinsic bioluminescence appears in over half of all light-producing species.
Davis and colleagues determined that the oldest lineage of fishes with bioluminescence belong to the order Stomiiformes—a group including fishes like the dragonfish (Fig. 2C) and hatchetfish (Fig. 2D)—and determined the ability emerged around the late Jurassic/early Cretaceous period. The prevalence of bioluminescent evolution in the Cretaceous could point to this ability being crucial in spurts of species diversity during those eras.

Big Picture
Constructing phylogenies like this may be time consuming and require significant computing power, but such studies provide great insight into the historical evolution of lineages. This helps inform future work examining macroevolution, or evolution at the level of families instead of individual species. There will always be an element of uncertainty, but as techniques improve and the gene databases expand, scientists like Davis and his colleagues should have a much easier time teasing apart the evolutionary history of unique adaptations.
I am a former PhD student from the University of Rhode Island, having discovered my love of teaching and informal science education in part through OceanBites! Since departing academia, I’ve focused on creating educational content for visitors at the New England Aquarium, Chincoteague Bay Field Station, and now the National Aquarium. I’ve also dabbled in co-creating a comedy/brainstorming podcast, ThunkTink, and enjoy getting lost in nature with my dogs.