Modern medicine is a fantastic thing; it’s improved people’s quality of life, helped curb the spread of disease, and provided numerous therapies over the last century. That does not mean it is without risk, or that the treatments doctors have developed will always stay effective. Some antibiotics that were effective decades ago are coming head to head with bacteria that have donned armor, so to speak, evolving or obtaining resistances to the medications designed to wipe them out. We can slow the development of antibiotic resistance (ABR) of bacteria in our bodies by practicing good hygiene (e.g. wash your hands) to lower the chances of getting sick, prescribing the correct medication from the beginning, and always taking the whole course of an antibiotic so infections are fully stopped. This is only part of the equation, though. We are connected to our environment, and no matter how hard we try, antibiotics or microbes resistant to them will enter the larger ecosystem.
ABR can emerge in two main ways. The first is through good old genetic mutation. While our bodies largely take up doses of antibiotics, small remainders pass through into our wastewater system. Filtration of wastewater by bacteria at a treatment facility exposes those microbes to low doses of the drugs. The dosage may not be enough to kill all the bacteria, but this stressor ensures genetically resistant bacteria will survive and reproduce more effectively. Once some of the bacteria are resistant, they can pass it directly along to daughter cells or release their resistant genes into the environment. In the latter case, neighboring colonies can take this genetic material up via horizontal gene transfer, thereby gaining resistance. As these newly resistant bacteria and their genes churn through the treatment facility’s system, some can make it through all filtration measures and end up in the effluent stream—that is, the water that leaves the facility and is reintroduced into the environment.
Just how much ABR organic material makes it through though, and how serious a problem is it if ABR genes are out there in the wild? In 2019, the US Center for Disease Control estimated about 2.8 million people contracted ABR infections per year—a number that will likely increase, especially if infection hotspots show up in areas many people visit. To get a better idea of how prevalent ABR genes were near the popular waters of Miami Beach, an interagency group (US Geological Survey, NOAA, Florida Dept. of Environmental Protection, and Broward County researchers) set off to find out, collecting water and sediment samples from three treatment plants north the city.
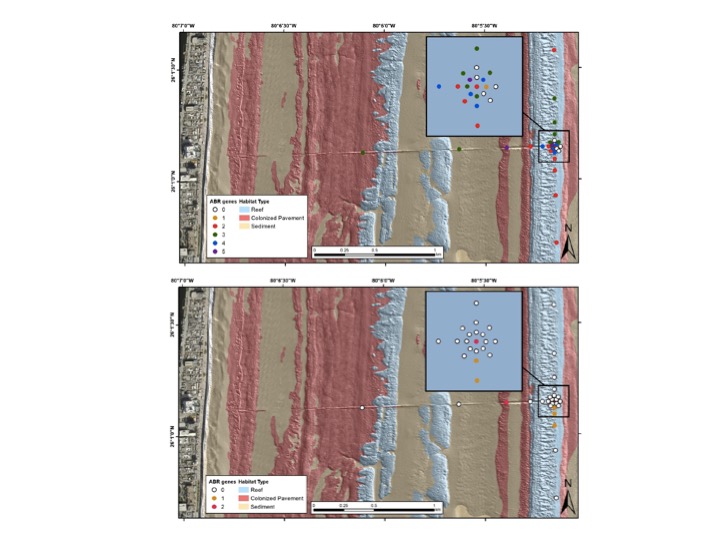
Incoming water to the facilities was sterilely sampled and SCUBA divers were deployed to collect water and sediment specimens from along and around the outfall pipes, located 3 km (~1.9 miles) offshore. Since Florida experiences two distinct seasons (a wet and a dry lasting between May-October and November-April, respectively), complete sets of samples from each season were collected (Fig. 2). Samples were then kept cold and shipped to the US Geological Survey lab for processing and analysis. DNA from the environmental samples was extracted and run through PCR (polymerase chain reaction). Using known primer sequences of 15 distinct ABR genes, any ABR DNA present was broken down, rebuilt, and copied repeatedly, allowing the team to isolate and identify which specific resistant sequences were floating around.
The wet season saw 94 detections (17 detections in 6 water samples, 77 detections in 32 sediment samples) of 9 ABR genes. Some genetic remnants were found only in water (e.g. tetB, an ABR gene that resists tetracycline, a drug used to treat everything from respiratory infections to acne) while others (e.g. aadA2, resistant against streptomycin, the medicine used to treat tuberculosis) were only found in sediment. This contrasted heavily with the dry season, where 79 detections representing 10 ABR genes appeared, with 68 detections noted from within 10 water samples, while only 11 detections pinged from sediment samples.
Based on the number of detections from the inflow pipes entering the facilities, the team saw an increase between the wet (summer) and dry (winter) seasons—ABR gene detections went from 11 to 46. This could coincide with an increase in people getting sick and taking medication over the winter months. The sharp drop in sediment ABR detections (77 from wet to 11 during dry) could suggest a larger environmental role, the team concluded. The wet season coincides with hurricane season, greater storm activity in general, and warmer water temperatures; these factors could contribute to increased mixing of sediment, its dispersal further south via coastal currents, and keeping bacteria at an optimal temperature for maximum reproduction.
The presence of these ABR genes does not mean there are colonies of superbugs lying in wait offshore, but it should be of concern. Because of mechanisms like horizontal gene transfer, it is possible new bacteria could gain resistance to medications they’ve never before encountered. In the event those genes spread throughout microbial colonies and humans become susceptible to the bacterial strain, we may be facing a tougher battle as our arsenal of treatments slowly shrinks. There are still ways we can mitigate the emergence of antibiotic resistance, and hopefully, future technologies will develop alongside that will help slow that spread even more.
I am a former PhD student from the University of Rhode Island, having discovered my love of teaching and informal science education in part through OceanBites! Since departing academia, I’ve focused on creating educational content for visitors at the New England Aquarium, Chincoteague Bay Field Station, and now the National Aquarium. I’ve also dabbled in co-creating a comedy/brainstorming podcast, ThunkTink, and enjoy getting lost in nature with my dogs.